Data Hierarchy
Each execution of the ASCENT pipeline requires a Run JavaScript
Object Notation (JSON) configuration file (<run_index>.json) that
contains indices for a user-defined set of JSON files. Specifically, a
JSON file is defined for each hierarchical domain of information: (1)
Sample: for processing segmented two-dimensional transverse
cross-sectional geometry of a nerve sample, (2) Model (COMSOL
parameters): for defining and solving three-dimensional FEM, including
geometry of nerve, cuff, and medium, spatial discretization (i.e.,
mesh), materials, boundary conditions, and physics, and (3) Sim
(NEURON parameters): for defining fiber models, stimulation waveforms,
amplitudes, and durations, intracellular test pulses (for example, when
seeking to determine block thresholds), parameters for the bisection search
protocol and termination criteria for thresholds, and flags to save
state variables. These configurations are organized hierarchically such
that Sample does not depend on Model or Sim, and
Model does not depend on Sim; thus, changes in Sim do
not require changes in Model or Sample, and changes in
Model do not require changes in Sample.
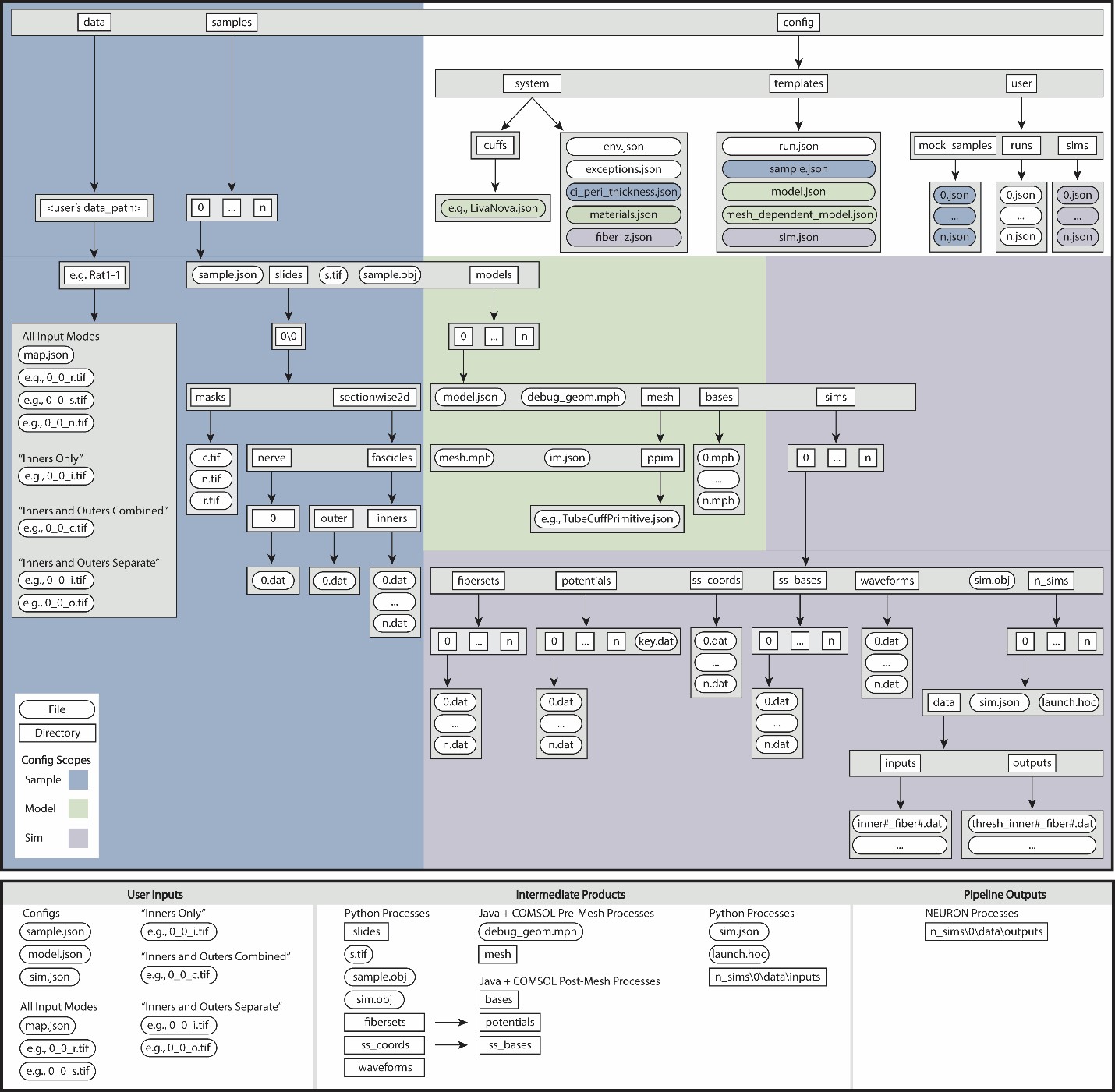
ASCENT (v1.0.0) pipeline file structure in the context of Sample (blue), Model (green), and Sim (purple) configurations. JSON Overview describes the JSON configuration files and their contents, and JSON Parameters details the syntax and data types of the key-value parameter pairs.
Batching and sweeping of parameters
ASCENT enables the user to batch rapidly simulations to sweep cuff electrode placement on the nerve, material properties, stimulation parameters, and fiber types. The first process of ASCENT prepares ready-to-submit NEURON simulations to model response of fibers to extracellular stimulation. The second process of ASCENT uses Python to batch NEURON jobs to a personal computer or compute cluster to simulate fiber response to extracellular stimulation. Each task submitted to a CPU simulates the response of a single fiber to either a set of finite amplitudes or a bisection search for threshold of activation or block, therefore creating an “embarrassingly parallel” workload.
Groups of fibers from the same Sample, Model, Sim,
waveform, contact weight (i.e., "src_weights" in Sim), and
fiberset (i.e., a group of fibers with the same geometry and channels
and occupy different (x,y)-locations in the nerve cross-section) are
organized in the same n_sim/ directory.
A Run creates simulations for a single Sample and all pairs
of listed Model(s) and Sim(s). A user can pass a list of
Run configurations in a single system call with "python run pipeline <run_indices>" to simulate multiple Sample
configurations in the same system call.
Sample and Model cannot take lists of parameters. Rather, if the user would like to assess the impact of ranges of parameters for Sample or Model, they must create additional Sample and Model configuration files for each parameter value.
Sim can contain lists of parameters in "active_srcs" (i.e., cuff
electrode contact weightings), "fibers", "waveform", and
"supersampled_bases".