JSON Configuration File Overview
We store parameters in JSON configuration files because the JSON format is accessible, readable, and well-documented for metadata interchange. The configuration files inform the pipeline in its operations and provide a traceable history of the parameters used to generate data.
For each JSON file, we provide a brief overview, a statement of where
the file must be placed in the directory structure, and a description of
its contents. For a detailed description of each JSON file (i.e., which parameters are required or optional, known value data types, and known values), see JSON Parameters. Though JSON does not allow comments, users may want to add
notes to a JSON file (e.g., to remember what a Sample,
Model, or Sim file was used to accomplish). The user can
simply add a key to the JSON file that is not in use (e.g., “notes”) and
provide a value (String) with a message.
Note
Example configuration files are provided in the config/templates/ directory.
User configuration files
run.json
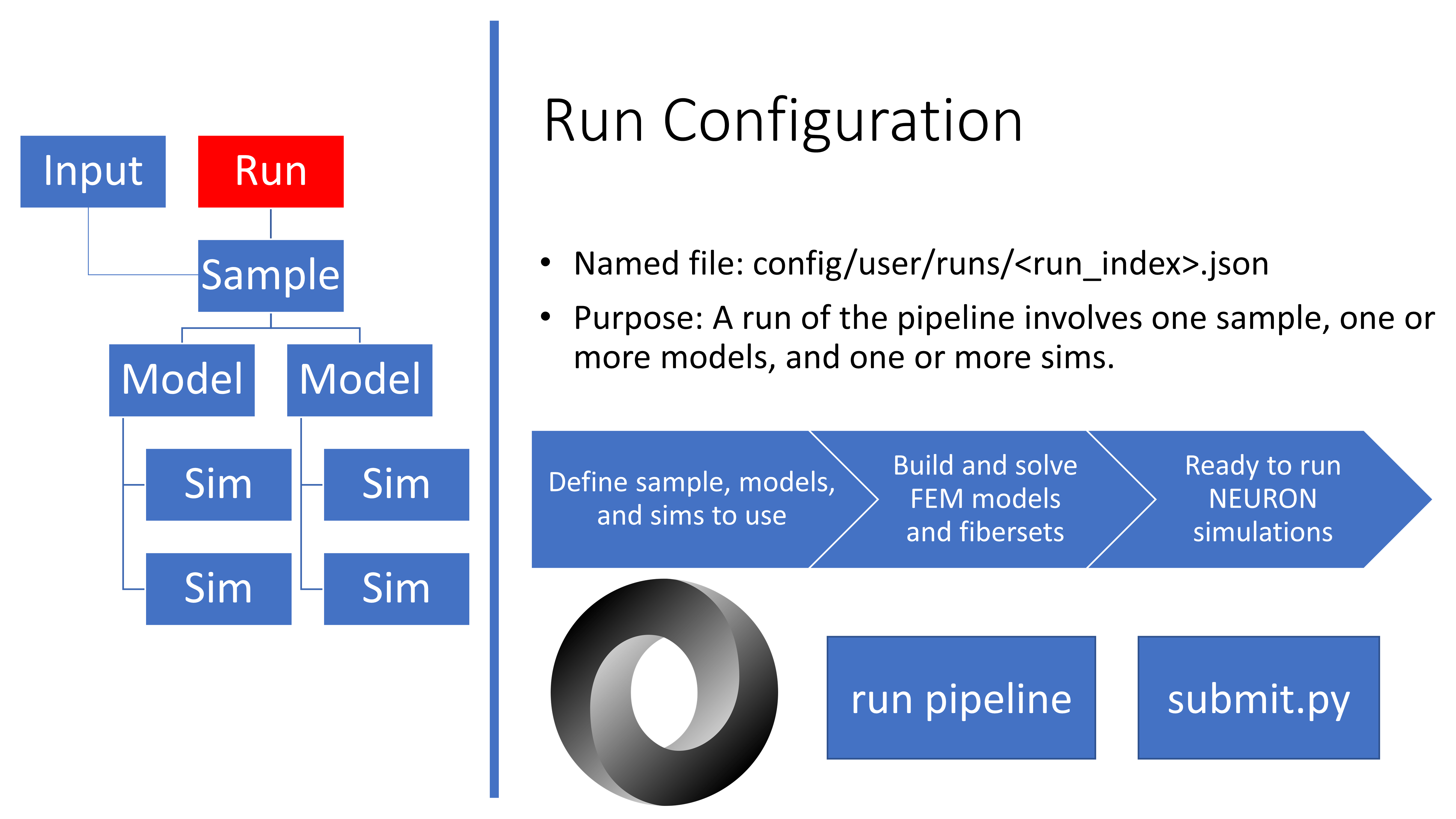
Overview of the run.json file
The run.json file is passed to pipeline.py to instruct the program
which Sample, Model(s), and Sim(s) to run. All
run.json files are stored in the config/user/runs/ directory.
Since each Sample, Model, and Sim is indexed, their
indices are used as the identifying values in run.json.
Additionally, the file contains break points that enable the user to
terminate the pipeline at intermediate processes, flags for user control
over which COMSOL files are saved, flags to selectively build a
"debug_geom.mph" file of just the nerve or cuff electrode, and flags
to recycle previous meshes where appropriate. Lastly, the run.json
file reports which Model indices were generated successfully in
COMSOL and indicates if the user is submitting the NEURON jobs to a
SLURM cluster or locally.
sample.json
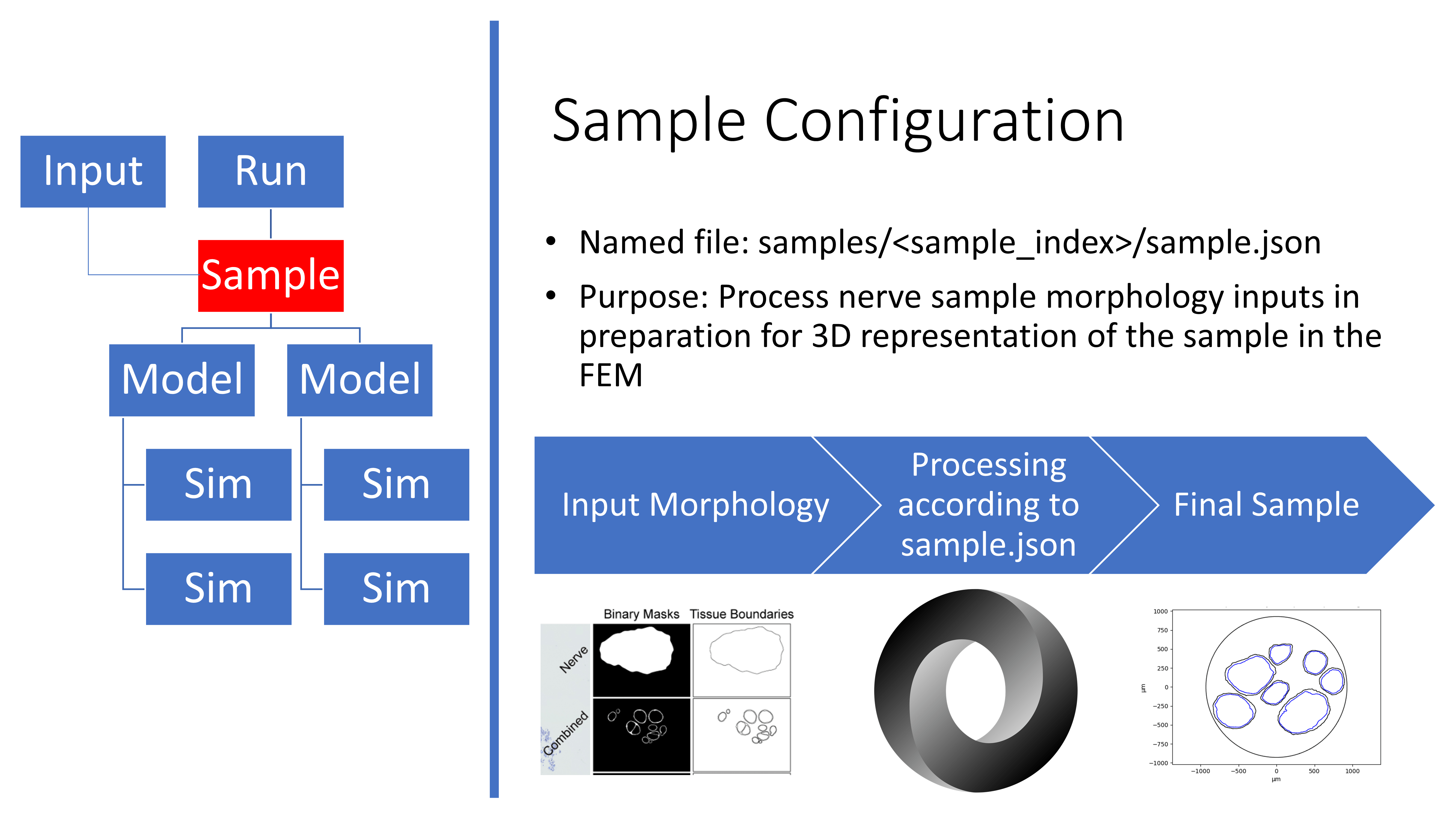
Overview of the sample.json file
An example Sample configuration file is stored in
config/templates/ for users to reference when defining their own
input nerve morphology from histology or from the mock nerve morphology
generator (Mock Morphology). A user’s sample.json file is saved in the
samples/<sample_index>/ directory. The file contains information
about the sample’s properties and how to process the nerve morphology
before placing the nerve in a cuff electrode. The pipeline’s processes
informed by Sample output the Python object sample.obj.
The information in Sample must parallel how the sample morphology
binary images are saved on the user’s machine (sample name, path to the
sample). The user must
define the "mask_input" parameter in Sample to indicate which set
of input masks will be used. Sample also contains parameters that
determine how the morphology is processed before being represented in
the FEM such as shrinkage correction (“shrinkage”), required minimum
fascicle separation ("boundary_separation"), presence of epineurium
(“nerve”), perineurium thickness ("ci_perineurium_thickness"),
deformation method (“deform”), reshaped nerve profile
("reshape_nerve"), and parameters for specifying CAD geometry input
file formats for nerve morphology (“write”). Sample also contains
information about the mask scaling (i.e.
"scale_bar_length" if supplying a binary image of a scale bar or, if no scale bar image, "scale_ratio").
The value of the "ci_perineurium_thickness" in Sample refers to a
JSON Object in config/system/ci_peri_thickness.json that contains
coefficients for linear relationships between inner diameter and
perineurium thickness (i.e., thkperi,inner =
a*(diameterinner) + b). In ci_peri_thickness.json, we
provided a "PerineuriumThicknessMode" named "GRINBERG_2008", which
defines perineurium thickness as 3% of inner diameter [Grinberg et al., 2008], and
relationships for human, pig, and rat vagus nerve perineurium thickness
(i.e., "HUMAN_VN_INHOUSE_200601", "PIG_VN_INHOUSE_200523", and
"RAT_VN_INHOUSE_200601") [Pelot et al., 2020]. As additional vagus nerve
morphometry data become available, users may define perineurium
thickness with new models by adding the coefficients to this JSON
file._**
model.json
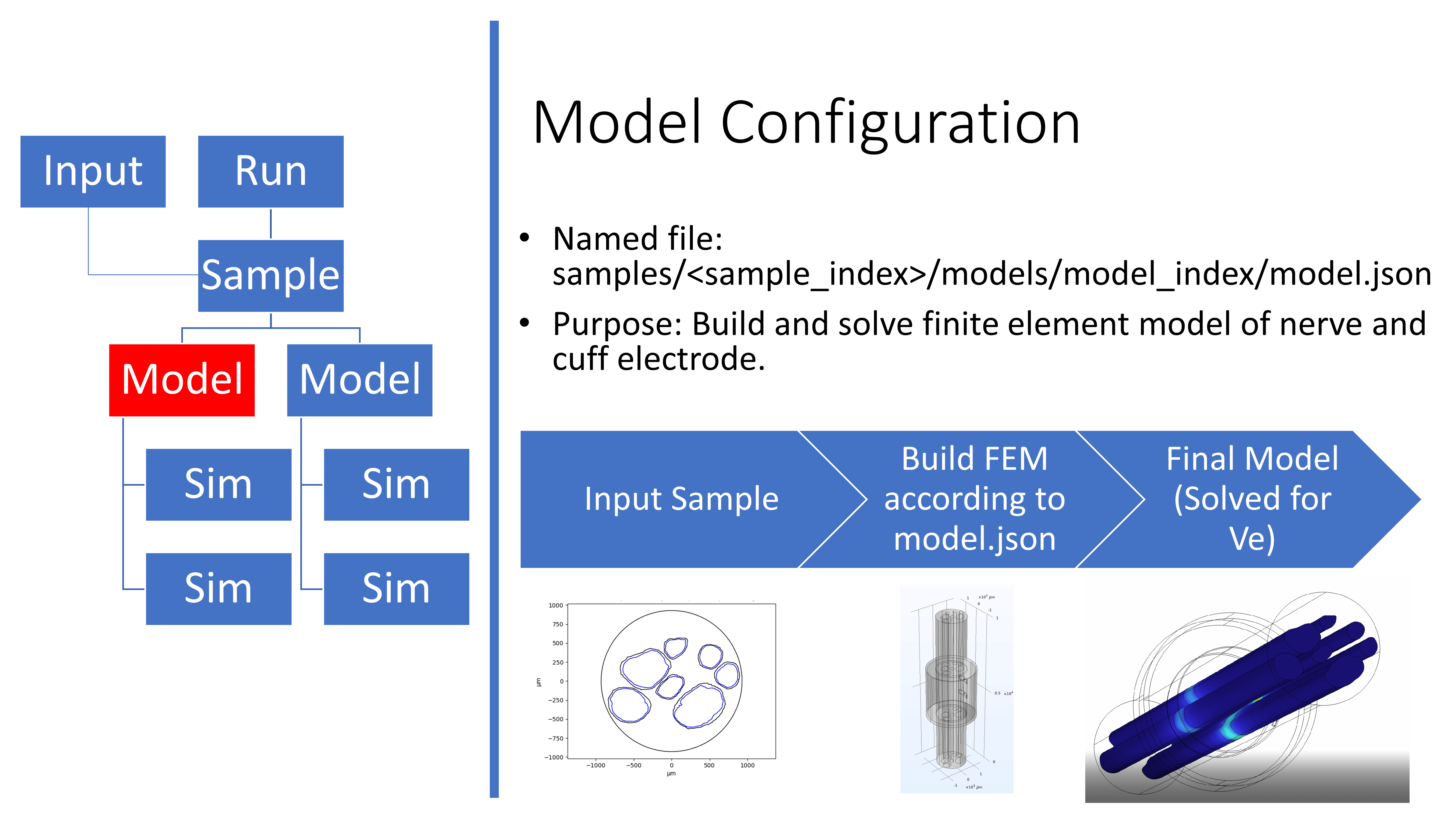
Overview of the model.json file
An example Model configuration file is stored in
config/templates/ for users to reference when creating their own
FEMs. As such, model.json, which is stored in the file structure in
samples/<sample_index>/models/<model_index>/, contains
information to define an FEM uniquely. Model defines the cuff
electrode geometry and positioning, the simulated environment (e.g.,
surrounding medium dimensions, material properties (including
temperature and frequency factors of material conductivity), and
physics), the meshing parameters (i.e., how the volume is discretized),
and output statistics (e.g., time required to mesh, mesh element quality
measures).
sim.json
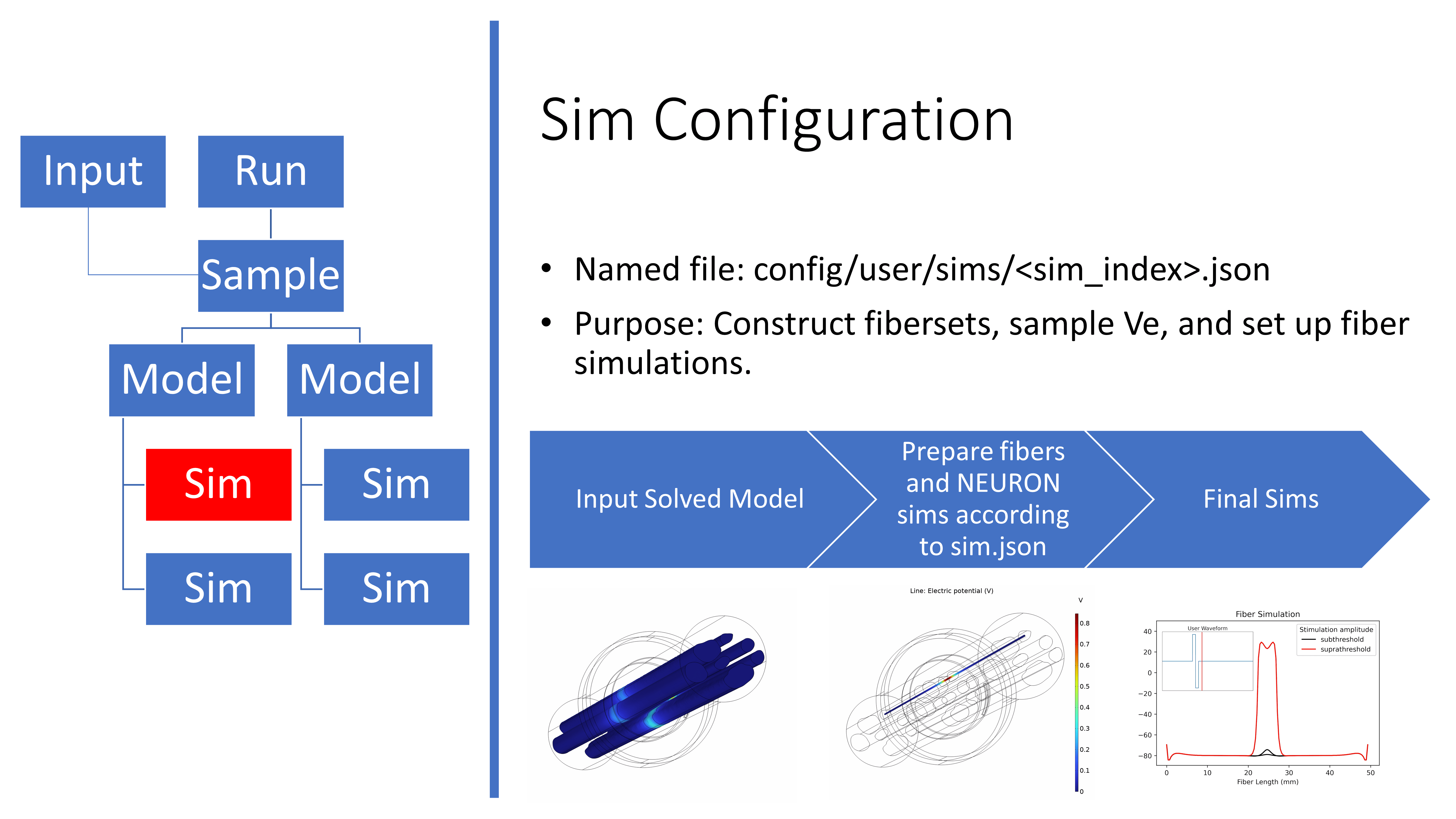
Overview of the sim.json file
An example Sim configuration file is stored in
config/templates/ for users to reference when creating their own
simulations of fiber responses to stimulation for a sample in a FEM. All
simulation configuration files are stored in the config/user/sims/
directory. Sim defines fiber types, fiber locations in the FEM,
fiber length, extracellular (e.g., pulse repetition frequency) and
intracellular stimulation, and input parameters to NEURON (e.g.,
parameters to be saved in the output, bisection search algorithm bounds and
resolution). Since users may want to sweep parameters at the Sim
configuration level (e.g., fiber types, fiber locations, waveforms), a
pared down copy of Sim that contains a single value for each
parameter (rather than a list) is saved within the corresponding
n_sims/ directory (Sim Parameters). These pared down files are provided for convenience,
so that the user can inspect which parameters were used in a single
NEURON simulation, and they do not hold any other function within the
pipeline.
Special use configuration files
mock_sample.json
The mock_sample.json file, which is stored in the file structure in
config/user/mock_samples/<mock_sample_index>/, is used to
define binary segmented images that serve as inputs to the pipeline. In
the “populate” JSON Object, the user must define the “PopulateMode”
(e.g., EXPLICIT, TRUNCNORM, UNIFORM defined by the “mode” parameter),
which defines the process by which the nerve morphology is defined in
the MockSample Python class. Each "PopulateMode" requires a certain set
of parameters to define the nerve and to define and place the fascicles;
the set of parameters for each "PopulateMode" are defined in
config/templates/mock_sample_params_all_modes.json.
Probabilistic “PopulateModes” (i.e., TRUNCNORM, UNIFORM) populate an
elliptical nerve with elliptical fascicles of diameter and eccentricity
defined by a statistical distribution. Since the nerve morphology
parameters are defined probabilistically, a “seed” parameter is required
for the random number generator to enable reproducibility. The fascicles
are placed at randomly chosen locations
within the nerve using a disk point picking method; the fascicles are
placed at a rotational orientation randomly chosen from
0-360o. If a fascicle is placed in the nerve without
maintaining a user-defined "min_fascicle_separation" distance from the
other fascicles and the nerve, another randomly chosen point within the
nerve is chosen until either a location that preserves a minimum
separation is achieved or the program exceeds a maximum number of
attempts ("max_attempt_iter").
The EXPLICIT "PopulateMode" populates an elliptical nerve with
elliptical fascicles of user-defined sizes, locations, and rotations.
The program validates that the defined fascicle ellipses are at least
"min_fascicle_separation" distance apart; otherwise, if the
conditions are not met, the program throws an error.
query_criteria.json
In data analysis, summary, and plotting, the user needs to inform the
program which output data are of interest. The query_criteria.json
file stores the “criteria” for a user’s search through previously
processed data. The query_criteria.json file may be used to guide
the Query class’s searching algorithm in the run() method. We suggest
that all query_criteria.json-like files are stored in the
config/user/query_criteria/ directory; however, the location of
these files is arbitrary, and when initializing the Query object, the
user must manually pass in the path of either the
query_criteria.json-like file or the hard-coded criteria as a
Python dictionary. An instance of the Query class contains the
“criteria” and an empty _result, which is populated by Query’s
run() method with found indices of Sample, Model, and
Sim that match the criteria given.
Query’s run() method loops through all provided indices (i.e.,
Sample, Model, Sim) in the query criteria, and calls
_match() when a possible match is found. Note that the presence of an
underscore in the _match() method name indicates that it is for
internal use only (not to be called by external files). The _match()
method compares its two inputs, (1) query_criteria.json and (2)
either Sample (i.e., sample.json), Model (i.e.,
model.json), or Sim (sim.json); the two JSON files are
loaded into memory as Python dictionaries. The method returns a Boolean
indicating if the input configuration file satisfies the restricted
parameter values defined in query_criteria.json. The user may
explicitly specify the indices of the Sample, Model, and Sim
configuration files of interest simultaneously with restricted criteria
for specific parameter values. The indices added will be returned in
addition to matches found from the search criteria in the Sample,
Model, and Sim criteria JSON Objects.
The query_criteria.json file contains JSON Objects for each of the
Sample, Model, and Sim configuration files. Within each JSON
Object, parameter keys can be added with a desired value that must be
matched in a query result. If the parameter of interest is found nested
within a JSON Object structure or list in the configuration file, the
same hierarchy must be replicated in the query_criteria.json file.
The query_criteria.json parameter "partial_matches" is a Boolean
indicating whether the search should return indices of Sample,
Model, and Sim configuration files that are a partial match, i.e.,
the parameters in query_criteria.json are satisfied by a subset of
parameters listed in the found JSON configuration.
The query_criteria.json parameter "include_downstream" is a
Boolean indicating whether the search should return indices of
downstream (Sample>Model>Sim) configurations that
exist if match criteria are not provided for them. For example, if only
criteria for a Sample and Model are provided, Query will
return the indices of Sample and Model that match the
criteria. In addition, the indices of the Sims downstream of the
matches are included in the result if "include_downstream" is true
(since the user did not specify criteria for Sim). Otherwise, if
"include_downstream" is false, no Sim indices are returned.
System configuration files
Note: system configuration files are located in config/system/.
env.json
The env.json file stores the file paths for:
COMSOL
Java JDK
The project path (i.e., the path to the root of the ASCENT pipeline)
Destination directory for NEURON simulations to run (this could be the directory from which the user calls NEURON, or an intermediate directory from which the user will move the files to a computer cluster)
Destination directory for exported datasets created with scripts/build_dataset.py
When the pipeline is run, the key-value pairs are stored as environment variables so that they are globally accessible.
materials.json
The materials.json file contains default values for material
properties that can be assigned to each type of neural tissue, each
electrode material, the extraneural medium, and the medium between the
nerve and inner cuff surface. The materials are referenced by using
their labels in the “conductivities” JSON Object of Model.
fiber_z.json
The fiber_z.json file defines z-coordinates to be sampled along the
length of the FEM for different fiber types to be simulated in NEURON.
In some instances, the section lengths are a single fixed value. In
other instances, such as the MRG model [McIntyre et al., 2002], the section lengths are
defined for each fiber diameter in a discrete list. Section lengths can
also be a continuous function of a parameter, such as fiber diameter,
defined as a mathematical relationship in the form of a string to be
evaluated in Python. Additionally, the file contains instructions (e.g.,
flags) that corresponds to fiber-type specific operations in NEURON.
ci_perineurium_thickness.json
In the case of fascicles with exactly one inner perineurium trace for
each outer perineurium trace, to reduce the required computational
resources, the pipeline can represent the perineurium using a thin layer
approximation in COMSOL (Perineurium Properties). Specifically, if Model’s "use_ci"
parameter is true, the perineurium is modeled as a surface with a sheet
resistance (termed “contact impedance” in COMSOL) defined by the product
of the resistivity and thickness. The thickness is calculated as half of
the difference between the effective circular diameters of the outer and
inner perineurium traces. If each fascicle is only defined by a single
trace (rather than inner and outer perineurium traces), the user chooses
from a list of modes in Sample for assigning a perineurium
thickness (e.g., 3% of fascicle diameter [Grinberg et al., 2008],
"ci_perineurium_thickness" parameter in Sample).
mesh_dependent_model.json
Since meshing can take substantial time and RAM, if the FEM has the same
geometry and mesh parameters as a previous model, this JSON file allows
the mesh to be reused if the mesh.mph file is saved. In
mesh_dependent_model.json, the keys match those found in Model,
however, instead of containing parameter values, each key’s value is a
Boolean indicating true if the parameter value must match between two
Model configurations to recycle a mesh, or false if a different
parameter value would not prohibit a mesh be reused. The
mesh_dependent_model.json file is used by our ModelSearcher Java
utility class (Java Utility Classes).