How to use ASCENT
Running the ASCENT Pipeline
See Running the Pipeline for instructions on how to run ASCENT.
The first three stages of ASCENT are invoked by the command python run pipeline.
This performs the necessary operations to prepare simulations of nerve stimulation.
The final stage of ASCENT is invoked by the command python submit.py, which runs the simulations.
After the simulations are run, the Query Class can be used to obtain the resultant data,
and the Plotter Module can be used to visualize the results.
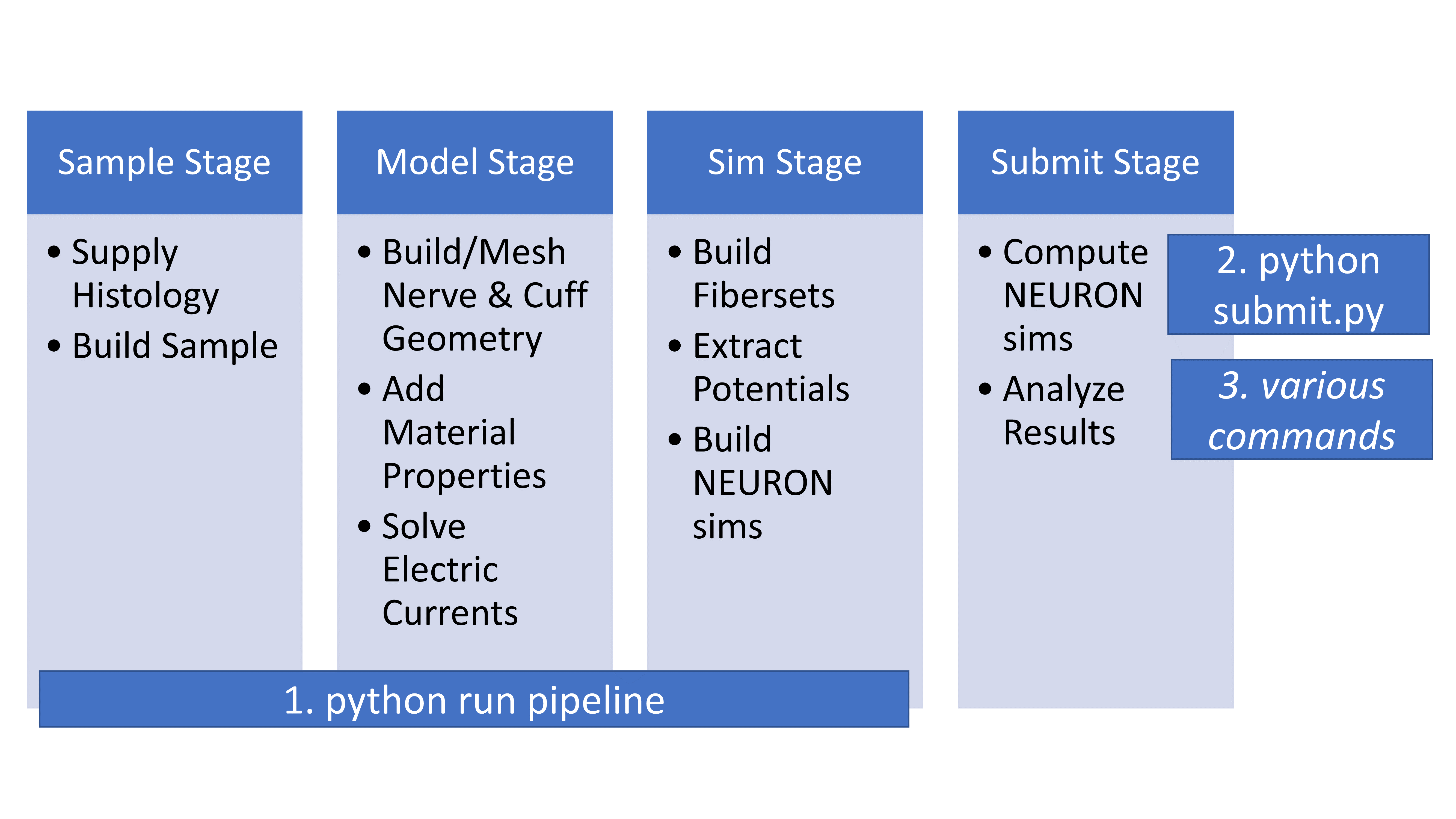
The stages of the ASCENT pipeline.
Submitting NEURON jobs
We provide scripts for the user to submit NEURON jobs in src/neuron/.
The submit.py script takes the input of the Run
configuration and submits a NEURON call for each independent fiber
within an n_sim/. These scripts are called using similar syntax as
pipeline.py: "./submit.<ext> <run indices>," where <run indices> is a space-separated list of integers. Note that these
submission scripts expect to be called from a directory with the
structure generated by Simulation.export_nsims() at the location
defined by "ASCENT_NSIM_EXPORT_PATH" in env.json.
Cluster submissions
When using a high-performance computing cluster running SLURM:
Set your default parameters, particularly your partition (can be found in
"config/system/slurm_params.json"). The partition you set here (default is “common” ) will apply to allsubmit.pyruns.submit.pywill run in cluster mode if it detects that"sbatch"is an available command. To override this behavior manually, use Command-Line Arguments.Many parameters which
submit.pysources from JSON configuration files can be overridden with command line arguments. For more information, see Command-Line Arguments.
Other Scripts
We provide scripts to help users efficiently manage data created by ASCENT. Run all of these scripts from the directory
to which you installed ASCENT (i.e., "ASCENT_PROJECT_PATH" as defined in config/system/env.json). For specific controls associated with each script, see Command Line Arguments.
scripts/import_n_sims.py
To import NEURON simulation outputs (e.g., thresholds, recordings of Vm) from your ASCENT_NSIM_EXPORT_PATH
(i.e., <"ASCENT_NSIM_EXPORT_PATH" as defined in env.json>/n_sims/<concatenated indices as sample_model_sim_nsim>/data/outputs/) into the native file structure
(see ASCENT Data Hierarchy, samples/<sample_index>/models/<model_index>/sims/<sim_index>/n_sims/<n_sim_index>/data/outputs/)
run this script from your "ASCENT_PROJECT_PATH".
python run import_n_sims <list of run indices>
The script will load each listed Run configuration file from config/user/runs/<run_index>.json to determine for which
Sample, Model(s), and Sim(s) to import the NEURON simulation data. This script will check for any missing thresholds and skip that import if any are found. Override this by passing the flag --force. To delete n_sim folders from your output directory after importing, pass the flag --delete-nsims. For more information, see Command-Line Arguments.
scripts/clean_samples.py
If you would like to remove all contents for a single sample (i.e., samples/<sample_index>/) EXCEPT a list of files
(e.g., sample.json, model.json) or EXCEPT a certain file format (e.g., all files ending .mph), use this script.
Run this script from your "ASCENT_PROJECT_PATH". Files to keep are specified within the python script.
python run clean_samples <list of sample indices>
scripts/tidy_samples.py
If you would like to remove ONLY CERTAIN FILES for a single sample (i.e., samples/<sample_index>/), use this script.
This script is useful for removing logs, runtimes, special,
and _.bat or _.sh scripts. Run this script from your "ASCENT_PROJECT_PATH". Files to remove are specified within the python script.
python run tidy_samples <list of sample indices>
Data analysis tools
Python Query class
The general usage of Query is as follows:
In the context of a Python script, the user specifies the search criteria (think of these as “keywords” that filter your data) in the form of a JSON configuration file (see
query_criteria.jsonin Query Parameters).These search criteria are used to construct a Query object, and the search for matching Sample, Model, and Sim configurations is performed using the method
run().The search results are in the form of a hierarchy of Sample, Model, and Sim indices, which can be accessed using the
summary()method.
Using this “summary” of results, the user is then able to use various convenience methods provided by the Query class to build paths to arbitrary points in the data file structure as well as load saved Python objects (e.g., Sample and Simulation class instances).
The Query class’s initializer takes one argument: a dictionary in the
appropriate structure for query criteria (Query Parameters) or a string value containing
the path (relative to the pipeline repository root) to the desired JSON
configuration with the criteria. Put concisely, a user may filter
results either manually by using known indices or automatically by using
parameters as they would be found in the main configuration files. It is
extremely important to note that the Query class must be
initialized with the working directory set to the root of the pipeline
repository (i.e., sys.path.append(ASCENT_PROJECT_PATH) in your
script). Failure to set the working directory correctly will break the
initialization step of the Query class.
After initialization, the search can be performed by calling Query’s
run() method. This method recursively dives into the data file structure
of the pipeline searching for configurations (i.e., Sample,
Model, and/or Sim) that satisfy query_criteria.json. Once
run() has been called, the results can be fetched using the summary()
accessor method. In addition, the user may pass in a file path to
excel_output() to generate an Excel sheet summarizing the Query
results. Finally, use the threshold_data() method to return a DataFrame
of thresholds with identifying information.
Query also has methods for accessing configurations and Python objects
within the samples/ directory based on a list of Sample,
Model, or Sim indices. The build_path() method returns the
path of the configuration or object for the provided indices. Similarly,
the get_config() and get_object() methods return the configuration
dictionary or saved Python object (using the Pickle package),
respectively, for a list of configuration indices. These tools allow for
convenient looping through the data associated with search criteria.
Example uses of these Query
convenience methods are included in examples/analysis/.
plot_sample.pyplot_fiberset.pyplot_waveform.py
Specialized ASCENT plots
See Plotter Module for more info; example uses of this module
are provided in examples/analysis.
Video generation for NEURON state variables
In examples/analysis/ we provide a script, plot_video.py, that creates
an animation of saved state variables as a function of space and time
(e.g., transmembrane potentials, MRG gating parameters). The user can
plot n_sim data saved in a data/output/ folder by referencing indices
for Sample, Model, Sim, inner, fiber, and n_sim. The
user may save the animation as either a *.mp4 or *.gif file to the
data/output/ folder.
The plot_video.py script is useful for determining necessary simulation
durations (e.g., the time required for an action potential to propagate
the length of the fiber) to avoid running unnecessarily long
simulations. Furthermore, the script is useful for observing onset
response to kilohertz frequency block, which is important for
determining the appropriate duration of time to allow for the fiber
onset response to complete.
Users need to determine an appropriate number of points along the fiber to record state variables. Users have the option to either record state variables at all Nodes of Ranvier (myelinated fibers) or sections (unmyelinated fibers), or at discrete locations along the length of the fiber (Sim Parameters).